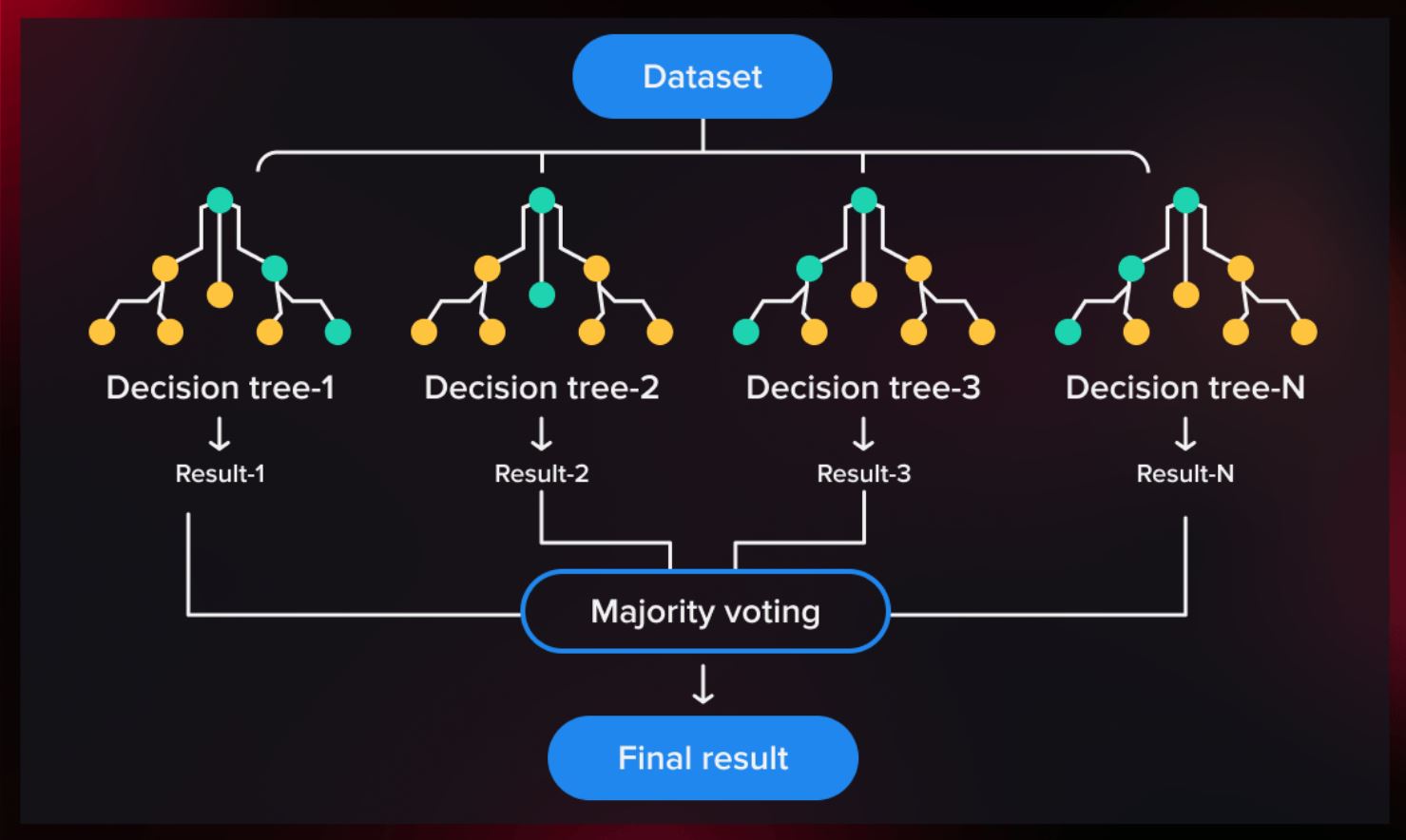
 Random Forest is a versatile machine learning tool that’s been used in healthcare, finance, agriculture, and more. It constructs a ‘forest’ of decision trees—each examining data through a sequence of questions, much like a flowchart. The final prediction is then made by combining the insights from each tree, similar to gathering opinions from several trusted experts.
Random Forest is a versatile machine learning tool that’s been used in healthcare, finance, agriculture, and more. It constructs a ‘forest’ of decision trees—each examining data through a sequence of questions, much like a flowchart. The final prediction is then made by combining the insights from each tree, similar to gathering opinions from several trusted experts.
If you’ve ever struggled with tuning your models, figuring out the perfect number of trees can seem daunting. Thankfully, the optRF package in R simplifies this process. In our guide, we walk you through using optRF alongside the ranger package not only to refine predictions but also to sharpen variable selection. We demonstrate this using SNPdata, a dataset featuring yield information from 250 wheat plants paired with 5000 genomic markers—a practical example if you’re interested in genomic prediction and breeding high-yield crops.
We start by splitting the dataset into training and testing sets. The opt_prediction function then helps determine an optimal tree count, recommending about 19,000 trees for making accurate predictions on unseen data. For assessing variable importance, the opt_importance function suggests ramping up to 40,000 trees. Increasing the tree count helps counteract the natural variability of Random Forest models, ensuring more stable results. However, more trees also demand more computation time, so it’s about finding the right balance.
The optRF package addresses this issue by analysing the relationship between the number of trees and model stability. Its plot_stability function provides a clear visual guide, showing that for our dataset, 40,000 trees yield near-perfect stability without excessive computational cost. Use these insights as a practical tip in your own work—you might find that a little fine-tuning goes a long way toward reliable modelling.








